Poisson regression for #TidyTuesday counts of R package vignettes
By Julia Silge in rstats tidymodels
March 16, 2022
This is the latest in my series of
screencasts demonstrating how to use the
tidymodels packages. Our team recently released new versions of
parsnip and the
parsnip-adjacent packages for specialized models to CRAN, and this screencast shows how to use some of these models with the
#TidyTuesday dataset on R package vignettes. 📄
Here is the code I used in the video, for those who prefer reading instead of or in addition to video.
Explore data
Our modeling goal is to understand how the number of releases and number of vignettes are related for R packages.
library(tidyverse)
cran <- read_csv("https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2022/2022-03-15/cran.csv")
What does this data look like, for a few packages that I maintain?
cran %>% filter(package == "tidytext")
## # A tibble: 8 × 5
## package version date rnw rmd
## <chr> <chr> <chr> <dbl> <dbl>
## 1 tidytext 0.2.1 2019-06-13 23:16:09 UTC 0 4
## 2 tidytext 0.2.2 2019-07-27 18:53:23 UTC 0 4
## 3 tidytext 0.2.3 2020-03-04 03:57:15 UTC 0 4
## 4 tidytext 0.2.4 2020-04-17 15:48:56 UTC 0 4
## 5 tidytext 0.2.5 2020-07-11 19:37:44 UTC 0 4
## 6 tidytext 0.2.6 2020-09-20 17:33:49 UTC 0 3
## 7 tidytext 0.3.0 2021-01-05 17:36:28 UTC 0 3
## 8 tidytext 0.3.1 2021-04-10 17:45:23 UTC 0 3
cran %>% filter(package == "rsample")
## # A tibble: 9 × 5
## package version date rnw rmd
## <chr> <chr> <chr> <dbl> <dbl>
## 1 rsample 0.0.1 2017-07-07 20:15:16 UTC 0 2
## 2 rsample 0.0.2 2017-11-12 00:07:22 UTC 0 3
## 3 rsample 0.0.3 2018-11-20 12:14:16 UTC 0 7
## 4 rsample 0.0.4 2019-01-07 01:49:42 UTC 0 7
## 5 rsample 0.0.5 2019-07-12 21:46:43 UTC 0 8
## 6 rsample 0.0.6 2020-03-31 18:46:21 UTC 0 8
## 7 rsample 0.0.7 2020-06-04 02:44:05 UTC 0 8
## 8 rsample 0.0.8 2020-09-23 16:08:48 UTC 0 8
## 9 rsample 0.0.9 2021-02-17 03:46:08 UTC 0 6
Let’s create a summarized dataset that computes, for each package, the first release data, the number of releases, and the number of vignettes as of the most recent release.
vignette_counts <-
cran %>%
group_by(package) %>%
summarise(
release_date = first(date),
releases = n(),
vignettes = last(rnw) + last(rmd)
)
What proportion of packages have no vignettes?
mean(vignette_counts$vignettes < 1)
## [1] 0.6611048
A lot!! We can see this in a histogram as well:
vignette_counts %>%
ggplot(aes(vignettes)) +
geom_histogram(bins = 12) +
scale_x_continuous(trans = scales::pseudo_log_trans(base = 10))
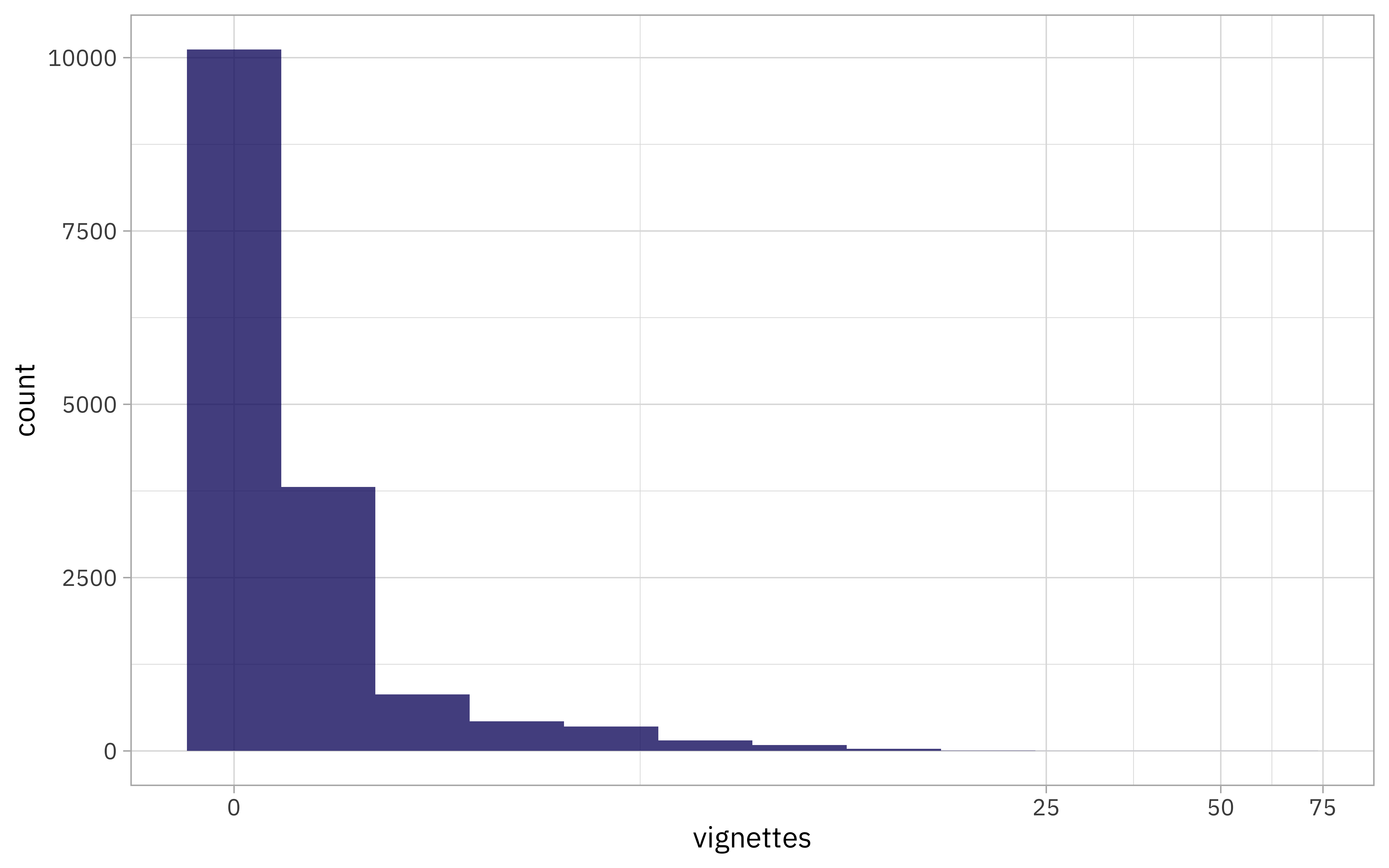
Just a few packages have a ton of vignettes:
vignette_counts %>% filter(vignettes > 20)
## # A tibble: 4 × 4
## package release_date releases vignettes
## <chr> <dttm> <int> <dbl>
## 1 catdata 2012-02-03 10:29:32 4 45
## 2 fastai 2020-11-05 18:26:03 9 24
## 3 HSAUR3 2014-04-17 10:31:57 11 23
## 4 pla 2015-08-18 21:58:53 2 61
Let’s make one more exploratory plot before creating models.
vignette_counts %>%
mutate(has_vignette = vignettes > 0) %>%
ggplot(aes(has_vignette, releases, color = has_vignette, fill = has_vignette)) +
geom_boxplot(size = 1.5, alpha = 0.2, show.legend = FALSE) +
scale_y_log10() +
coord_flip() +
labs(x = "Has a vignette?")
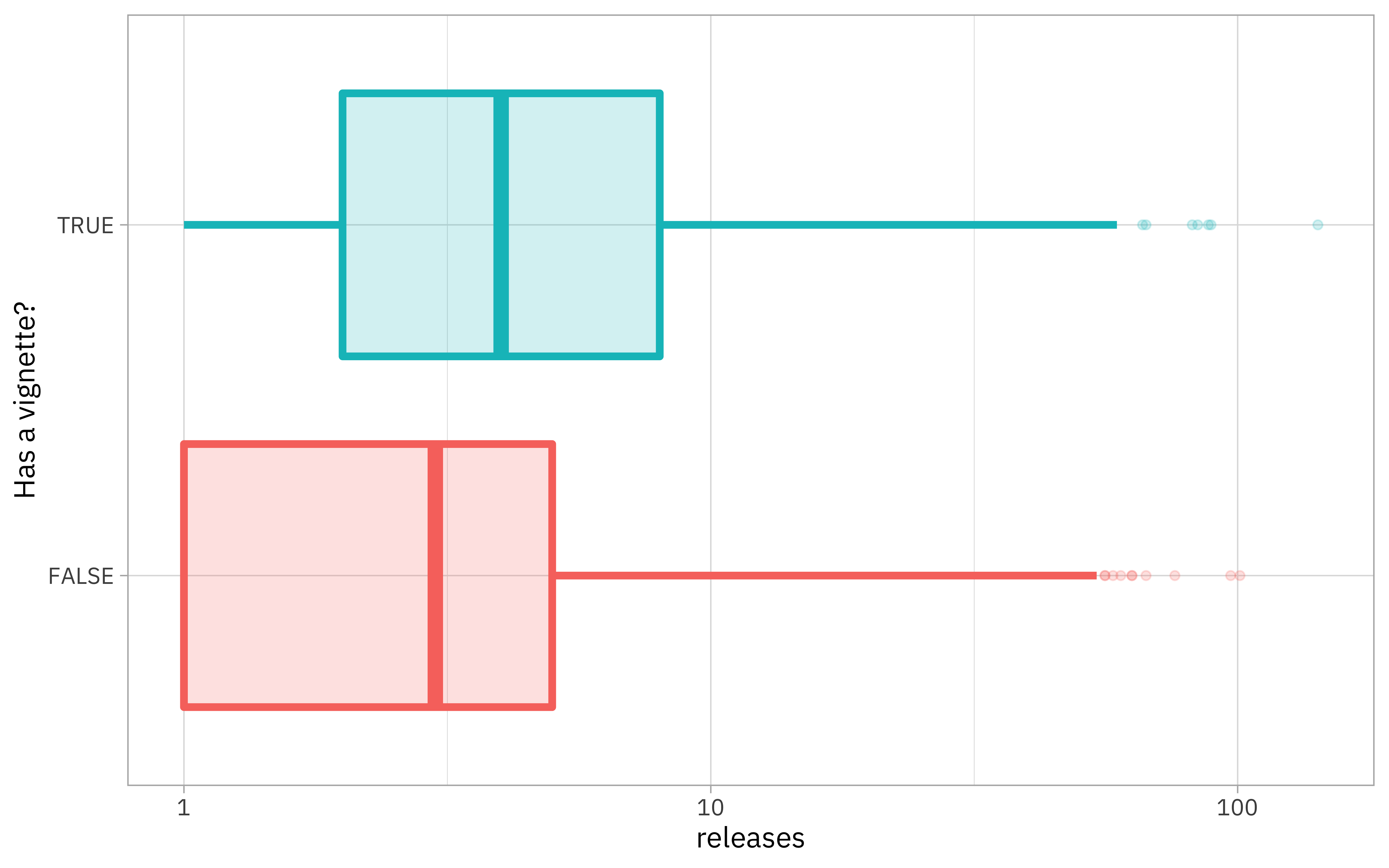
Looks like packages with more releases are more likely to have a vignette.
Using Poisson regression
We have a number of specialized packages, outside the core tidymodels packages, for less general, more specialized data analysis and modeling tasks. One of these is poissonreg, for Poisson regression models such as those we can use with this count data.
library(tidymodels)
library(poissonreg)
poisson_wf <- workflow(vignettes ~ releases, poisson_reg())
fit(poisson_wf, data = vignette_counts)
## ══ Workflow [trained] ══════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: poisson_reg()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## vignettes ~ releases
##
## ── Model ───────────────────────────────────────────────────────────────────────
##
## Call: stats::glm(formula = ..y ~ ., family = stats::poisson, data = data)
##
## Coefficients:
## (Intercept) releases
## -0.58265 0.03047
##
## Degrees of Freedom: 15793 Total (i.e. Null); 15792 Residual
## Null Deviance: 28880
## Residual Deviance: 27820 AIC: 41110
This model says that packages with more releases have more vignettes. Since poissonreg is not a core tidymodels package, we need to load it separately via library(poissonreg).
Zero-inflated Poisson: ZIP!!!
A better model for this dataset on R package vignettes might be zero-inflated Poisson, since there are so many zeroes. A ZIP model like this mixes two models, one that generates zeroes and one that models counts with the Poisson distribution. There are two sets of covariates for these two models, that can be different:
- one for the count data
- one for the probability of zeroes
How can we create this kind of model in tidymodels?
zip_spec <- poisson_reg() %>% set_engine("zeroinfl")
zip_wf <- workflow() %>%
add_variables(outcomes = vignettes, predictors = releases) %>%
add_model(zip_spec, formula = vignettes ~ releases | releases)
fit(zip_wf, data = vignette_counts)
## ══ Workflow [trained] ══════════════════════════════════════════════════════════
## Preprocessor: Variables
## Model: poisson_reg()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## Outcomes: vignettes
## Predictors: releases
##
## ── Model ───────────────────────────────────────────────────────────────────────
##
## Call:
## pscl::zeroinfl(formula = vignettes ~ releases | releases, data = data)
##
## Count model coefficients (poisson with log link):
## (Intercept) releases
## 0.1679 0.0247
##
## Zero-inflation model coefficients (binomial with logit link):
## (Intercept) releases
## 0.2019 -0.0271
The coefficients here are different than when we didn’t use a ZIP model, but we still see that packages with more releases have more vignettes (and packages with fewer releases are more likely to have zero vignettes).
Notice the formula argument we used in add_model(); this kind of
special model formula can be used with a lot of the parsnip-adjacent packages. The formula vignettes ~ releases | releases specifies which columns affect the counts and which affect the model for the probability of zero counts. Here these are the same, but
they don’t have to be.
Bootstrap intervals for the coefficients
You can do all the normal things with these models, depending on the purpose of your model. Often these kinds of models are trained to be used in inference, so let’s show how you might determine bootstrap intervals for model coefficients.
First let’s create a set of bootstrap resamples:
folds <- bootstraps(vignette_counts, times = 1000, apparent = TRUE)
folds
## # Bootstrap sampling with apparent sample
## # A tibble: 1,001 × 2
## splits id
## <list> <chr>
## 1 <split [15794/5840]> Bootstrap0001
## 2 <split [15794/5819]> Bootstrap0002
## 3 <split [15794/5830]> Bootstrap0003
## 4 <split [15794/5858]> Bootstrap0004
## 5 <split [15794/5860]> Bootstrap0005
## 6 <split [15794/5805]> Bootstrap0006
## 7 <split [15794/5875]> Bootstrap0007
## 8 <split [15794/5777]> Bootstrap0008
## 9 <split [15794/5845]> Bootstrap0009
## 10 <split [15794/5752]> Bootstrap0010
## # … with 991 more rows
Now let’s create a little function to get out the coefficients for the probability-of-zero-counts part of our ZIP model. (We could instead or in addition get out the Poisson/count part of the ZIP model.)
get_coefs <- function(x) {
x %>%
extract_fit_engine() %>%
tidy(type = "zero")
}
fit(zip_wf, data = vignette_counts) %>% get_coefs()
## # A tibble: 2 × 6
## term type estimate std.error statistic p.value
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) zero 0.202 0.0310 6.51 7.63e-11
## 2 releases zero -0.0271 0.00322 -8.41 3.94e-17
We can now take this function and use it for all of our bootstrap resamples with our ZIP model.
ctrl <- control_resamples(extract = get_coefs)
doParallel::registerDoParallel()
set.seed(123)
zip_res <- fit_resamples(zip_wf, folds, control = ctrl)
zip_res
## # Resampling results
## # Bootstrap sampling with apparent sample
## # A tibble: 1,001 × 5
## splits id .metrics .notes .extracts
## <list> <chr> <list> <list> <list>
## 1 <split [15794/15794]> Apparent <tibble [2 × 4]> <tibble> <tibble>
## 2 <split [15794/5840]> Bootstrap0001 <tibble [2 × 4]> <tibble> <tibble>
## 3 <split [15794/5819]> Bootstrap0002 <tibble [2 × 4]> <tibble> <tibble>
## 4 <split [15794/5830]> Bootstrap0003 <tibble [2 × 4]> <tibble> <tibble>
## 5 <split [15794/5858]> Bootstrap0004 <tibble [2 × 4]> <tibble> <tibble>
## 6 <split [15794/5860]> Bootstrap0005 <tibble [2 × 4]> <tibble> <tibble>
## 7 <split [15794/5805]> Bootstrap0006 <tibble [2 × 4]> <tibble> <tibble>
## 8 <split [15794/5875]> Bootstrap0007 <tibble [2 × 4]> <tibble> <tibble>
## 9 <split [15794/5777]> Bootstrap0008 <tibble [2 × 4]> <tibble> <tibble>
## 10 <split [15794/5845]> Bootstrap0009 <tibble [2 × 4]> <tibble> <tibble>
## # … with 991 more rows
What is in that .extracts column?
zip_res$.extracts[[33]]
## # A tibble: 1 × 2
## .extracts .config
## <list> <chr>
## 1 <tibble [2 × 6]> Preprocessor1_Model1
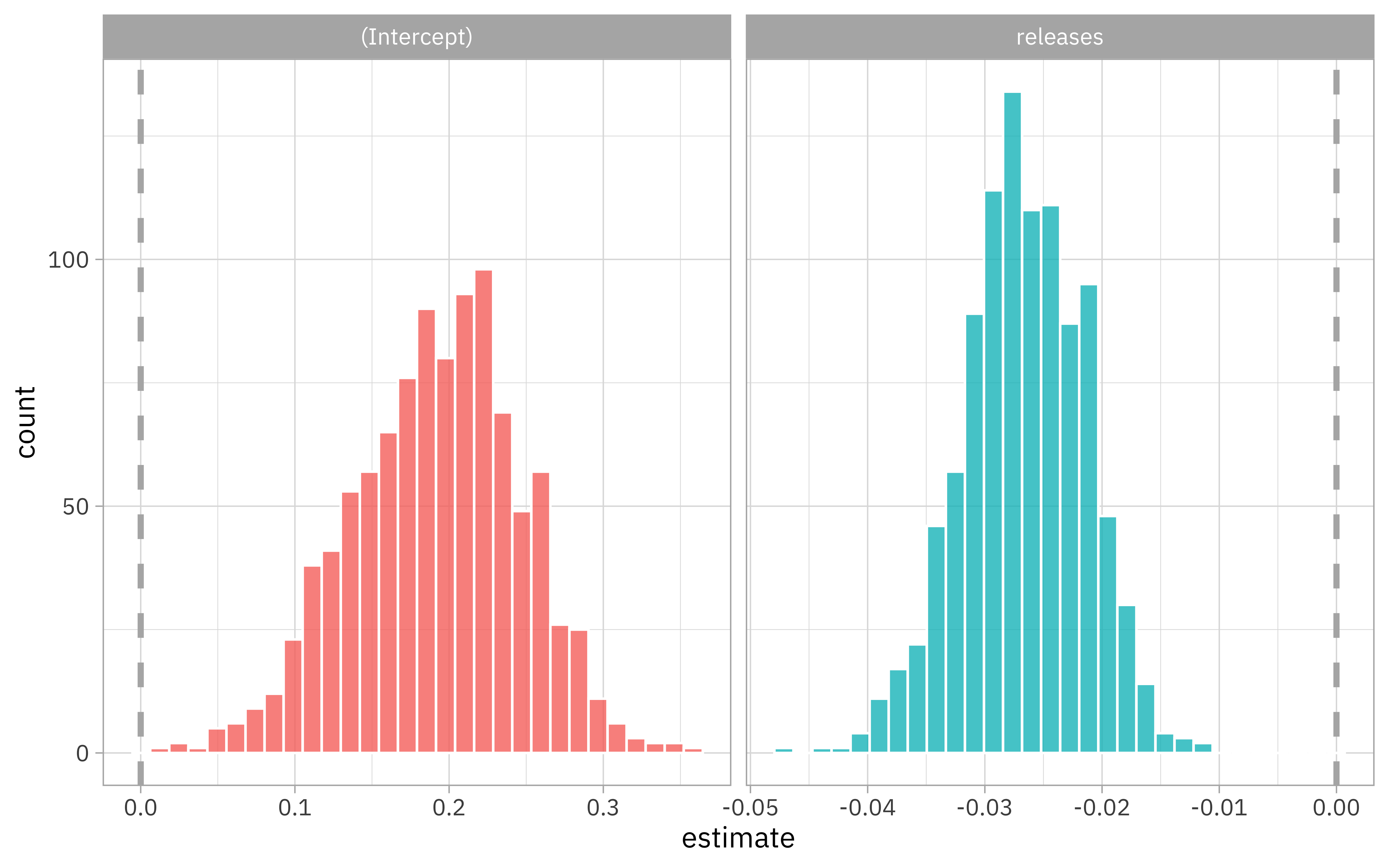
We can use tidyr to get those out, and then we can visualize the bootstrap intervals.
zip_res %>%
select(id, .extracts) %>%
unnest(.extracts) %>%
unnest(.extracts) %>%
ggplot(aes(x = estimate, fill = term)) +
geom_histogram(color = "white", alpha = 0.8, show.legend = FALSE) +
facet_wrap(~term, scales = "free_x") +
geom_vline(xintercept = 0, lty = 2, size = 1.2, color = "gray70")
- Posted on:
- March 16, 2022
- Length:
- 7 minute read, 1439 words
- Categories:
- rstats tidymodels
- Tags:
- rstats tidymodels